Meet Our Expert: Tony Person
“At the forefront of our mission, we are harnessing the power of cutting-edge SEC-MALS (Size Exclusion Chromatography with Multi-Angle Light Scattering) analysis to propel us into a position of leadership within the protein product industry. Our unwavering commitment to excellence drives us to continuously refine and expand our protein product portfolio, ensuring that we not only meet but exceed the highest industry standards.”
-Anthony Person Ph.D.
Vice President, Business Unit Leader- Proteins & Enzymes

Introduction
From clinical trials to basic science, recombinant proteins are critical for advancing your research. Bio-Techne understands that for researchers to maximize their success, our recombinant proteins must display rigorous quality control and consistency to provide you with a superior product you can trust. Beyond ensuring your recombinant proteins have high levels of biological activity across lots, we also utilize cutting-edge protein characterization techniques.
Protein characterization should have a minimal set of tests which include the following: purity, identity (molecular weight, MW), oligomeric state (monomer, dimer, trimer…) and homogeneity (aggregation state)1,2. SEC-MALS is an established method for precise and reliable quantification of protein molecular weight that can perform the above metrics and characterize proteins even further.
|
SEC-MALS |
Size Exclusion Chromatography-Multi-Angle Light Scattering |
|
MW |
Molecular Weight. |
|
SEC-HPLC |
Size Exclusion Chromatography-High Performance Liquid Chromatography |
|
Modifier |
Proteins are often conjugated with other materials (modifiers) like sugars (glycosylation) or PEG. |
|
Particle peak |
Particles may come from environment (dust) or column (column shedding). |
|
Aggregate |
Disordered or mis-folded proteins clump together forming an aggregate. Light Scattering is highly sensitive in detecting protein aggregation. |
|
Hydrodynamic volume |
SEC-HPLC separates molecules according to their size (hydrodynamic volume) and shape. |
|
Oligomeric states |
Oligomers are molecules that form from the interaction of two or more separate molecules (monomers). |
|
Monomer |
A molecule that is the basic unit for polymers, which are the building blocks of proteins. A monomer is a non-repeating structure. |
|
Dimer |
Oligomer made up of two monomers. |
|
Homodimer |
Oligomer made up of two identical monomers. |
| Trimer | Oligomer made up of three monomers. |
| Homotrimer | Oligomer made up of three identical monomers. |
|
Tetramer |
Oligomer made up of four monomers. |
|
Polydispersity |
Measure of the width of the molar mass distribution. Polydispersity is 1.000 for a uniform monodisperse population. |
|
Retention time |
The amount of time a protein spends on the column after it has been injected. |
|
Biclonal |
Appearance of two different monoclonal antibodies in sample. |
|
PTM |
Post-translational modification. |
| DLS | Dynamic Light Scattering |
What is SEC-MALS?
SEC-MALS is a technique that is best described as the sum of its parts. The first part of the technique consists of Size Exclusion Chromatography – High Performance Liquid Chromatography (SEC-HPLC), which allows sample fractionation via a column. The second part of the technique involves a Multi-Angle Light Scattering (MALS) detector, which determines the MW of separated fractions by using well-defined equations3,5 and concentration detectors (UV/RI).

Figure 1: Typical SEC-MALS Set Up. A SEC-MALS configuration typically involves having a HPLC/FPLC system with a SEC column, a UV detector, a MALS detector and a dRI detector. A sample is injected into the system and flows through the attached SEC column where the sample is separated. Once eluted from the column, the sample enters the UV detector, through the MALS detector and ultimately through the dRI detector. By simultaneously measuring the light scattering and the concentration of the molecule(s) as they pass through the detectors, the molar mass of the molecule(s) can be determined.4
SEC-MALS journey of a monomer, dimer and trimer

Figure 2: 1. A protein sample is run through chromatography to separate monomers, dimers and trimers by their hydrodynamic volume. 2. The molecules then pass through the UV detector, which provides composition data based on the molecules absorbance level to UV light. 3. Traveling through the MALS detector, molecules are hit with a laser. There are at least three detectors positioned to measure the light scattering of the laser off the sample to determine molecular weight. 4. The sample passes through the differential refractive index (dRI) detector, which measures the change in refractive index of the sample solution to a blank solvent to determine concentration. 5. The output of these detectors is summarized in a chromatograph. 6. Analysis of the chromatograph can show the separation of the monomers, dimers and trimers over time, as well as their absorbance and molar mass concentration.
Principles Behind SEC-MALS
First, the principle of SEC-HPLC is based on the separation of molecules through a stationary phase (column) based on the molecules hydrodynamic volume (size) and shape – not MW.
Second, the principle of MALS involves a laser beam of polarized light that is focused onto the sample molecule and the scattered light is detected at multiple angles (at least x3 angles are required). By utilizing the MALS detector along with two concentration detectors, the molar mass and weight-fraction of a modifier can be determined.
Highlighting peaks from a typical SEC-MALS profile

Figure 3: Understanding SEC-MALS analysis. (A) Example of a graph showing BSA run through SEC-MALS. (B) Zoom in of graph in A showing the separation of the monomer-dimer-trimer species of BSA using SEC-MALS.
There are a variety of methods available to characterize the molecular mass of proteins, which include the following: SEC-HPLC, SDS-PAGE, Native PAGE, Mass Spectrometry (MS), Analytical Ultracentrifugation (AUC), Capillary Electrophoresis (CE) and Light scattering (LS). Each of the above methods have pros and cons that go with them when determining molecular mass.5
SEC-MALS is a common method for characterizing the molecular mass of macromolecules; however, there are limitations that need to be considered (as described in the table below).
| Advantage | Limitations | |
| SEC-HPLC |
1. Easy, well established non-destructive method to measure masses of proteins based on hydrodynamic volumes and sizes. 2. Resolve different protein species: monomers/dimers/oligomers/fragments |
1. Need to use known calibration standard(s). 2. Assumes sample of interest has similar conformation (globular) as calibration standards. Not fit to measure elongated proteins. 3. Assume sample does not interact with column. Not fit to measure “sticky” proteins. |
| Light Scattering SLS (MALS) DLS (QELS) |
1. Assessing homogeneity of sample using non-destructive measurements. Light scattering is able to detect trace amounts of aggregates. 2. Column-Free measurements possible. |
1. Low resolution method that can’t separate molecules that are closely related (monomer/dimer). 2. Large aggregates, even a small amount, may affect measurement. |
| SEC-MALS |
1. Provides absolute molar mass independent of retention time, calibration standards, protein conformation and column interactions. 2. Non-destructive measurement where no protein modification is needed. 3. Protein conjugate analysis: [Glycosylation, Antibody-Drug conjugate (ADC)…] 4. Determination of the stoichiometry of protein/protein complexes. |
1. Identically sized molecules will not be separated. 2. Should only be used with highly pure samples containing well-resolved peaks to obtain accurate measurements. 3. Large complexes that may be susceptible to dissociation by dilution and/or shear forces during chromatographic separation, SEC-MALS may underestimate MW. 4. SEC-MALS typically requires extensive equilibration to obtain a baseline signal. |
At Bio-Techne, we apply SEC-MALS to a variety of applications

A. Proteins that are glycosylated and/or contain a Fc-tag are commonly sized incorrectly with SEC-HPLC and DLS. However, SEC-MALS has provided the correct MW of the protein.
| Sample | Cat# | Predicted MW (Monomer) | SDS-PAGE MW | SEC-HPLC MW | DLS MW | SEC-MALS MW |
| rhB7-H1/Fc | 156-B7 | 52 kDa | 140-150 kDa | 264 kDa | 265 kDa | 146 kDa |
| rhN-Cadherin/Fc/His | 1388-NC | 89 kDa | 230-260 kDa | 520 kDa | 605 kDa | 245 kDa |
B. Initial SEC-HPLC and DLS analysis of rhFAP/His provided a lower MW than anticipated, as if the protein was a monomer. However, the desired structure of the protein is a homodimer because the monomer is inactive. With the protein showing activity, it is unlikely a monomer. SEC-MALS identified protein as a homodimer.
| Sample | Cat# | Predicted MW (Monomer) | SDS-PAGE MW | SEC-HPLC MW | DLS MW | SEC-MALS MW |
| rhFAP/His | 3715-SE | 86 kDa | 85 kDa | 117 kDa | 130 kDa | 182 kDa |
C. When glycosylation (PTM’s) make it difficult to call a protein a monomer or a dimer using SEC-HPLC and DLS. From SEC-HPLC and DLS, this protein appears to be in range of a dimer based on Predicted MW. SEC-MALS identified the protein as a monomer.
| Sample | Cat# | Predicted MW (Monomer) | SDS-PAGE MW | SEC-HPLC MW | DLS MW | SEC-MALS MW |
| rhBTN/His | 8467-BT | 25 kDa | 36 kDa | 46 kDa | 46 kDa | 28 kDa |

Utilizing SEC-MALS, we are able to accurately assign oligomeric peaks (rhIL-10, 1064-ILB). The preferred oligomeric state is the Dimer.
| rhIL-10 | SEC-MALS MW | Oligomeric State | Polydispersity | Retention Time |
| Peak 1 | 22 kDa | Monomer | 1.001 | 19.2 - 19.5 min |
| Peak 2 | 37 kDa | Dimer | 1.002 | 17.9 - 18.5 min |
| Peak 3 | 79 kDa | Tetramer | 1.000 | 16.5 - 16.8 min |

By using SEC-MALS we are able to show that regardless of retention time these peaks are intact antibodies.
| Sample | SEC-MALS MW | Polydispersity | Retention Time |
| Antibody 1 (Red) | 149 kDa | 1.000 | 15.6 – 16.0 min |
| Antibody 2 (Blue) | 147 kDa | 1.000 | 17.3 - 18.0 min |

A group within Bio-Techne was having trouble obtaining consistent results with an outsourced mAb.
By SEC-MALS we observe at least x2 populations of antibodies, making this mAb at least biclonal and possibly explaining why they were observing inconsistencies in their assay.
| Vendor mAb | SEC-MALS MW | Polydispersity | Retention Time |
| Peak 1 | 160 kDa | 1.002 | 15.8 - 16.2 min |
| Peak 2 | 153 kDa | 1.003 | 16.4 - 17.1 min |

We utilize SEC-MALS to ensure the protein is in the correct structure (rhTNFa – 10291-TA). The correct structure is a homotrimer.
| rhTNFa | Predicted MW (Monomer) | SEC-MALS MW | Polydispersity | Retention Time |
| Peak 1 | 17 kDa | 52 kDa | 1.000 | 18.6 - 19.0 min |

By using SEC-MALS (protein conjugate analysis) we are able to show that the “higher” than expected MW is in fact due to glycosylation (rhGDNF, 212-GD).
| rhGDNF | Predicted MW (Monomer) | Total MW | Protein MW | Modifier MW (Glycosylation) |
| Peak 1 | 12 kDa | 32 kDa | 24 kDa | 8 kDa |
| GDNF is a disulfide-linked homodimer with x2 potential N-glycosylation sites. | ||||

We obtained competitor material to run on Bio-Techne SEC-MALS system to see how the SEC-MALS data analysis would compare. The MW difference (~10 kDa) is enough to cast doubt on competitor MW claim.
| Competitor MALS-MW | 54 kDa |
| BioTechne MALS-MW | 63 kDa |
Proteins Analyzed by SEC-MALS

| SEC-MALS Data | Result |
| Retention Time | 18.5-19.0 min |
| MW-Predicted (Monomer) | 17.0 kDa |
| MW-MALS | 53.1 kDa |
| Polydispersity | 1.000 |
| System Suitability: BSA Monomer 66.4 ± 3.32 kDa | Pass |
Recombinant Human TNF-alpha (Catalog # 210-TA) has a molecular weight (MW) of 53.1 kDa as analyzed by SEC-MALS, suggesting that this protein is a homotrimer. MW may differ from predicted MW due to post-translational modifications (PTMs) present (i.e. Glycosylation).
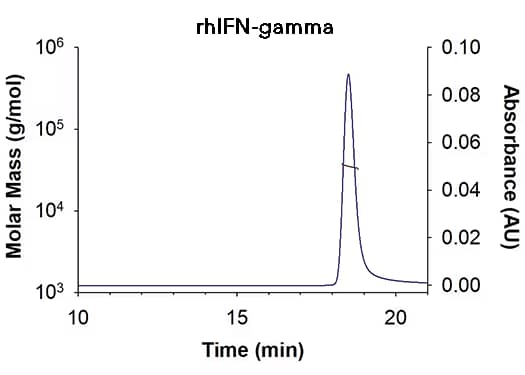
| SEC-MALS Data | Result |
| Retention Time | 18.3-18.8 min |
| MW-Predicted (Monomer) | 16.9 kDa |
| MW-MALS | 34.9 kDa |
| Polydispersity | 1.001 |
| System Suitability: BSA Monomer 66.4 ± 3.32 kDa | Pass |
Recombinant Human IFN-gamma (Catalog # 285-IF) has a molecular weight (MW) of 34.9 kDa as analyzed by SEC-MALS, suggesting that this protein is a homodimer. MW may differ from predicted MW due to post-translational modifications (PTMs) present (i.e. Glycosylation).

| SEC-MALS Data | Result |
| Retention Time | 16.0-16.6 min |
| MW-Predicted (Monomer) | 16.0 kDa |
| MW-MALS | 17.2 kDa |
| Polydispersity | 1.001 |
| System Suitability: BSA Monomer 66.4 ± 3.32 kDa | Pass |
Recombinant human FGF basic/FGF2/bFGF, 145 aa TC Grade (Catalog # 4114-TC) has a molecular weight (MW) of 17.2 kDa as analyzed by SEC-MALS, suggesting that this protein is a monomer. MW may differ from predicted MW due to post-translational modifications (PTMs) present (i.e. Glycosylation).
-
P4EU (2019) Protein Quality Standard PQS
-
Arbre Mobieu (2019) Guidelines on Protein Quality Control.
-
Shamir, M. et al. (2019) Characterization of Protein Oligomers by Multi-angle Light Scattering. Encyclopedia of Analytical Chemistry pp 1- 17.
-
D’Arcy et al. (2021) Understanding Absolute Stoichiometry of Oligomeric Protein Complexes Using SEC-MALS
-
Some, D. et al. (2019) Characterization of Proteins by Size-Exclusion Chromatography Coupled to Multi-Angle Light Scattering (SEC-MALS) J. Vis. Exp. 148:e59615, doi:10.3791/59615.






